How you can use cBioPortal
cBioPortal offers countless ways to explore and interpret cancer genomics data—without needing to code. Below, we walk through a series of research questions using KRAS as an example gene to show how to use public datasets to uncover meaningful insights.
📝 Note: These examples use the public cBioPortal instance but can be replicated on the Fred Hutch instance as well.
Example: Investigating KRAS in Cancer
Q1: How often is KRAS mutated in cancer?
- Visualize different mutations across colorectal, lung, and pancreatic cancers in this oncoprint → KRAS is mutated in 26% of patients.
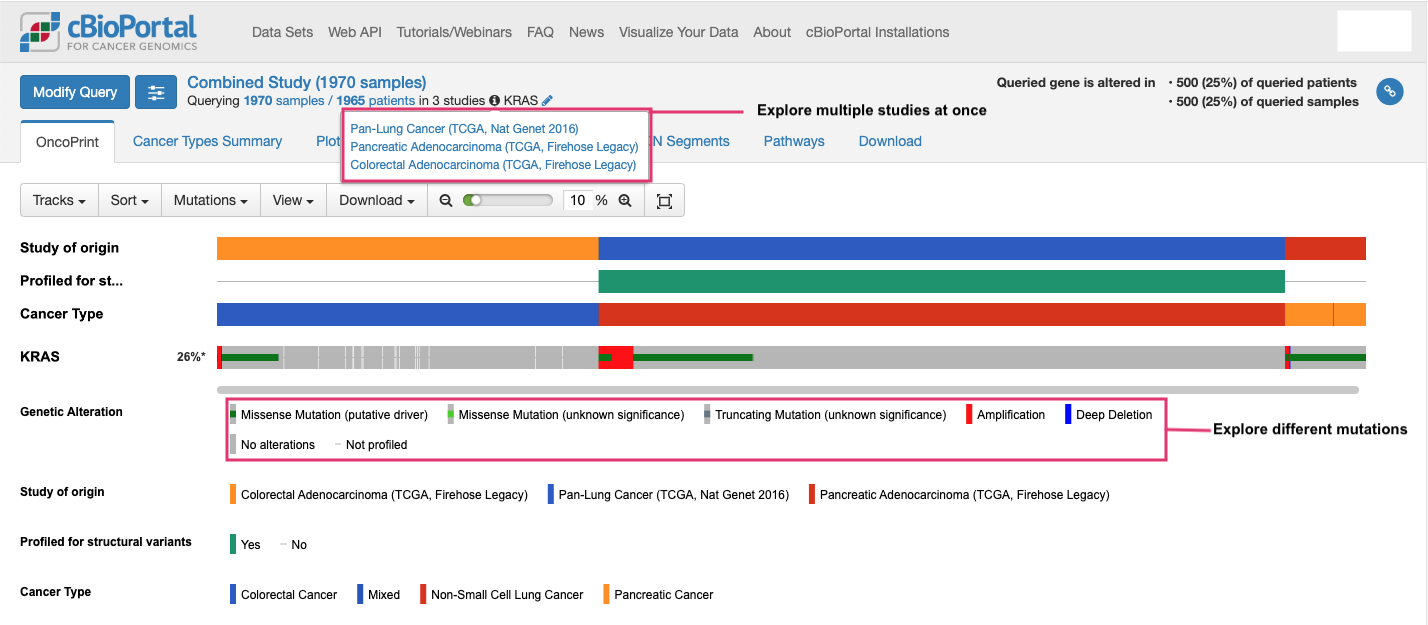
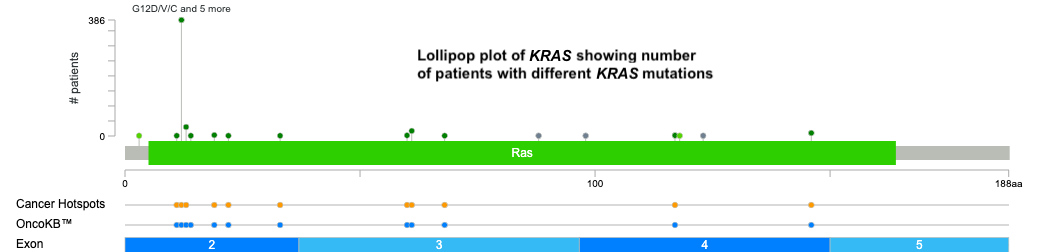
- Explore mutation types using lollipop plots → KRAS G12 position is mutated most frequently.
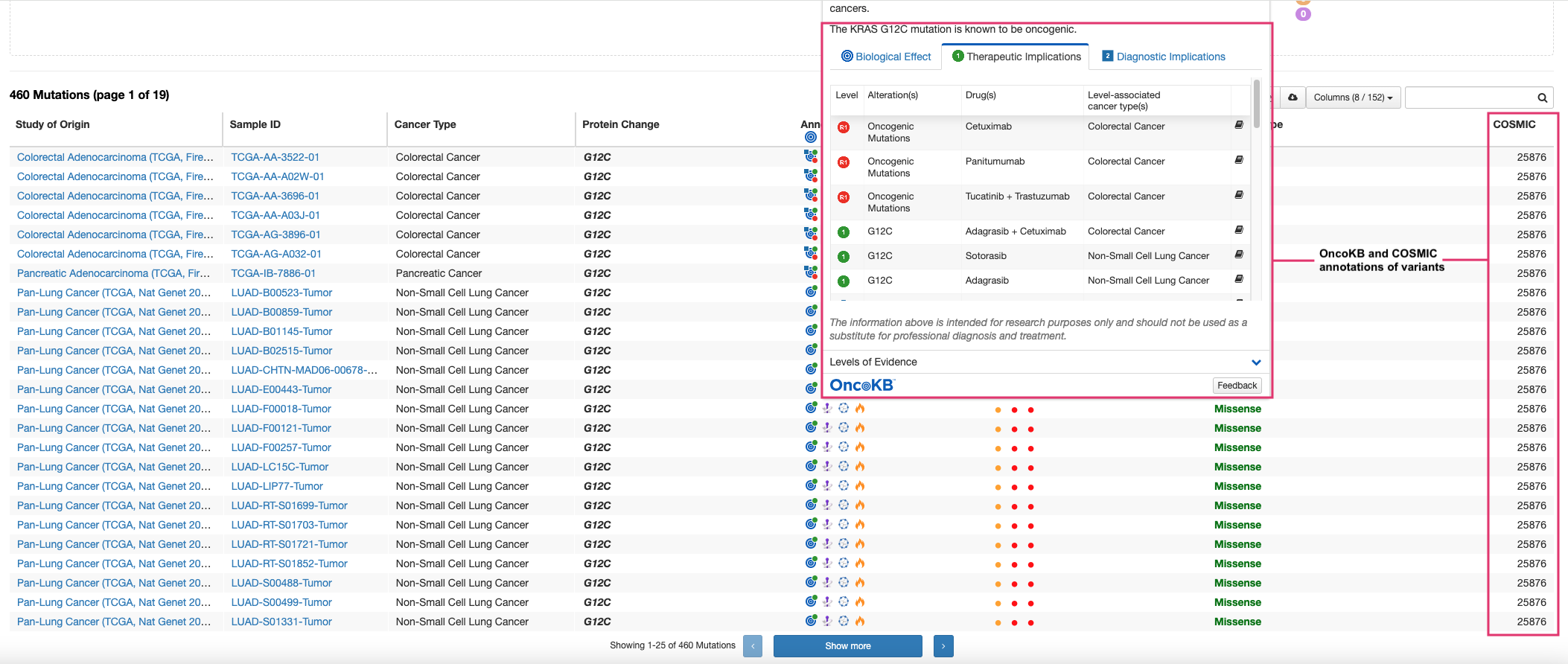
- Access OncoKB and COSMIC annotations for each variant → Most KRAS G12 mutations are oncogenic.
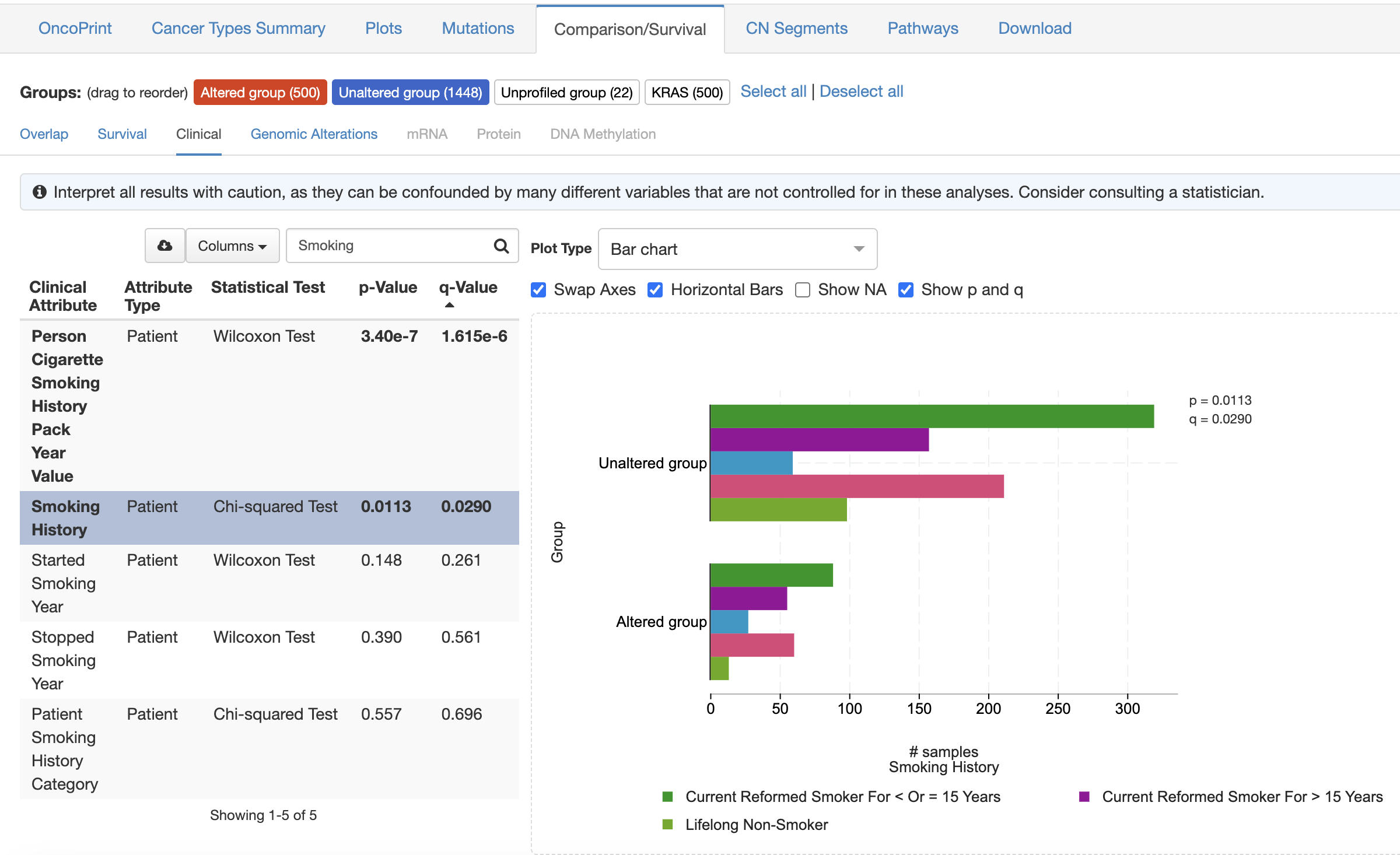
Q2: Are KRAS mutations associated with clinical factors?
- Add tracks for clinical correlates such as smoking history to an oncoprint to explore their relationship with genomic data → KRAS mutations occur more frequently in patients with a history of smoking.
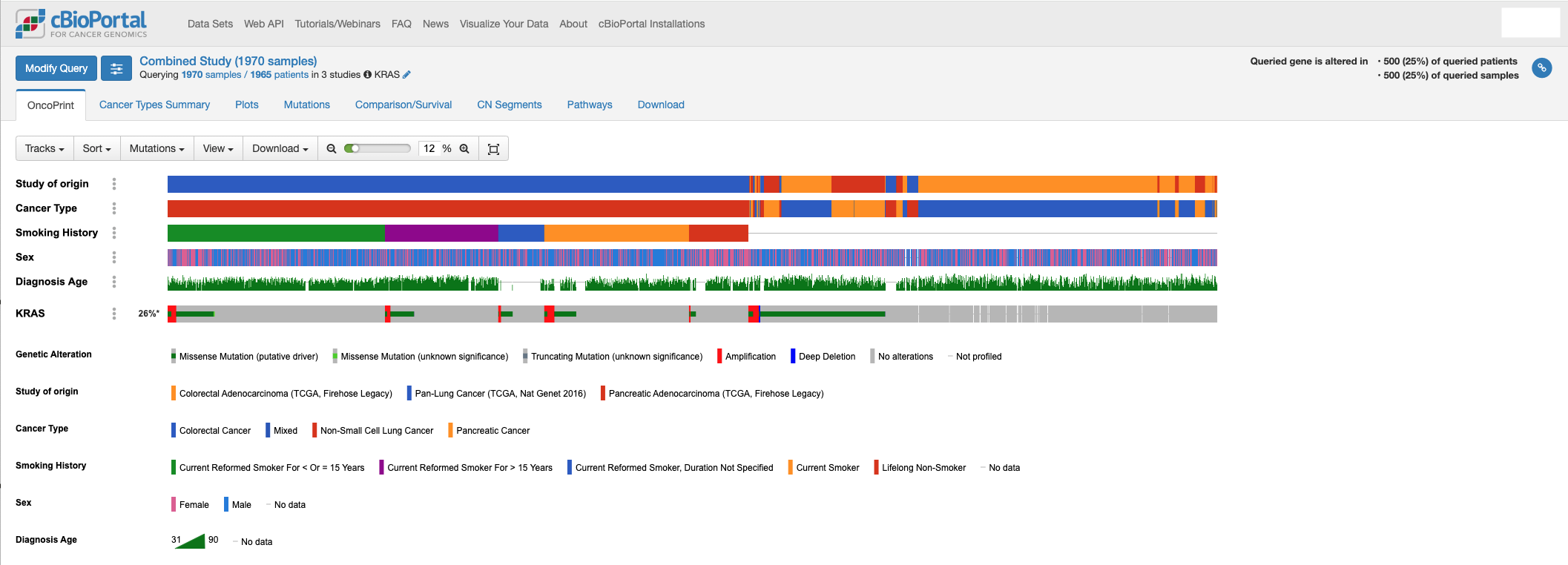
- Use the plots tab to correlate clinical variables with mutation status → Patients with a history of smoking have more KRAS mutations.
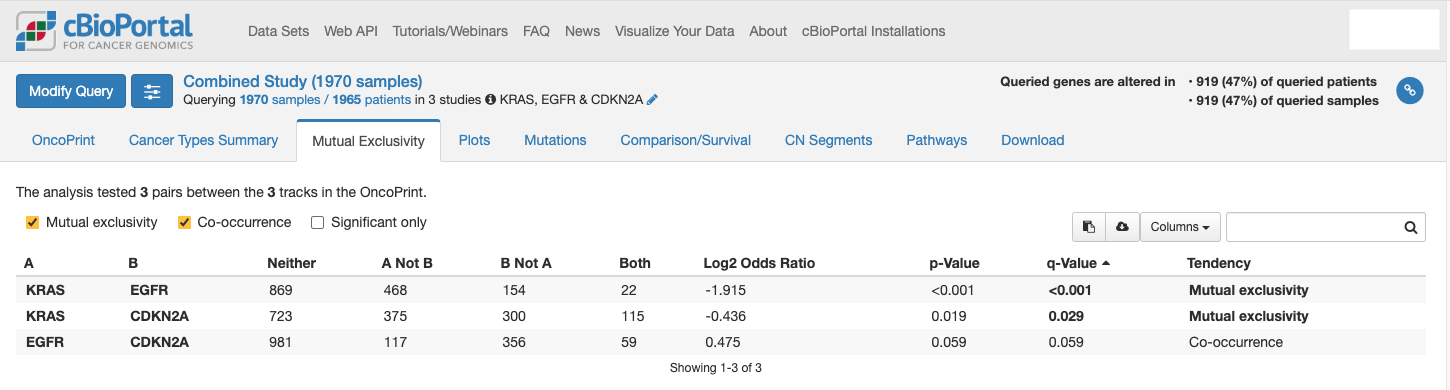
Q3: Do KRAS mutations co-occur with other mutations?
- Use the mutual exclusivity tool to examine co-mutations or exclusivity→ KRAS mutations are exclusive from other common driver gene mutations.
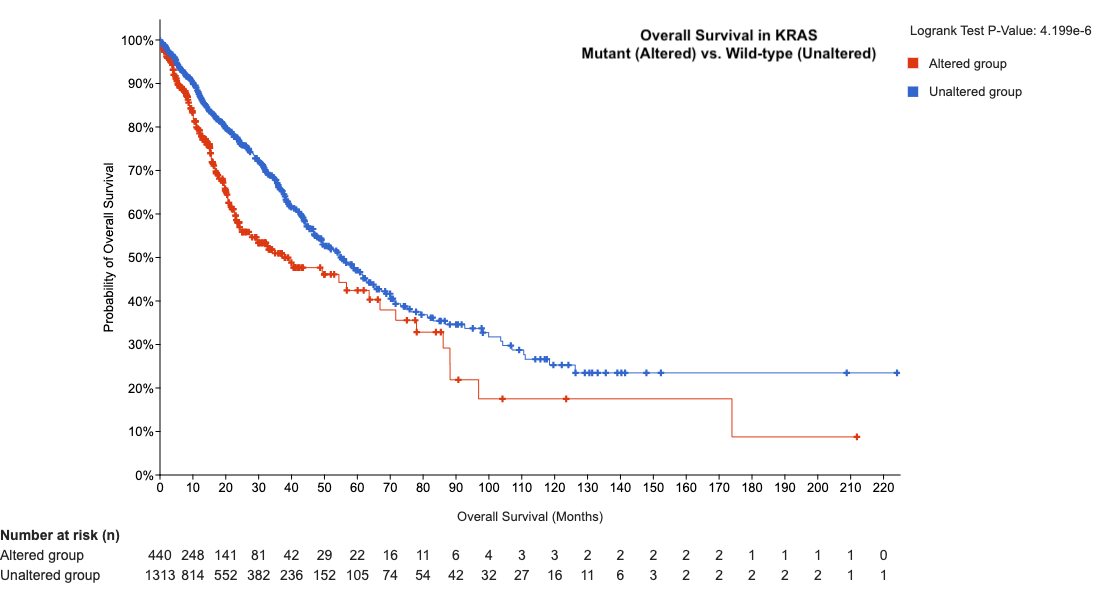
Q4: Do KRAS mutations affect survival?
- View overall survival differences between KRAS-mutant and wild-type tumors → Patients with mutant KRAS have lower overall survival.
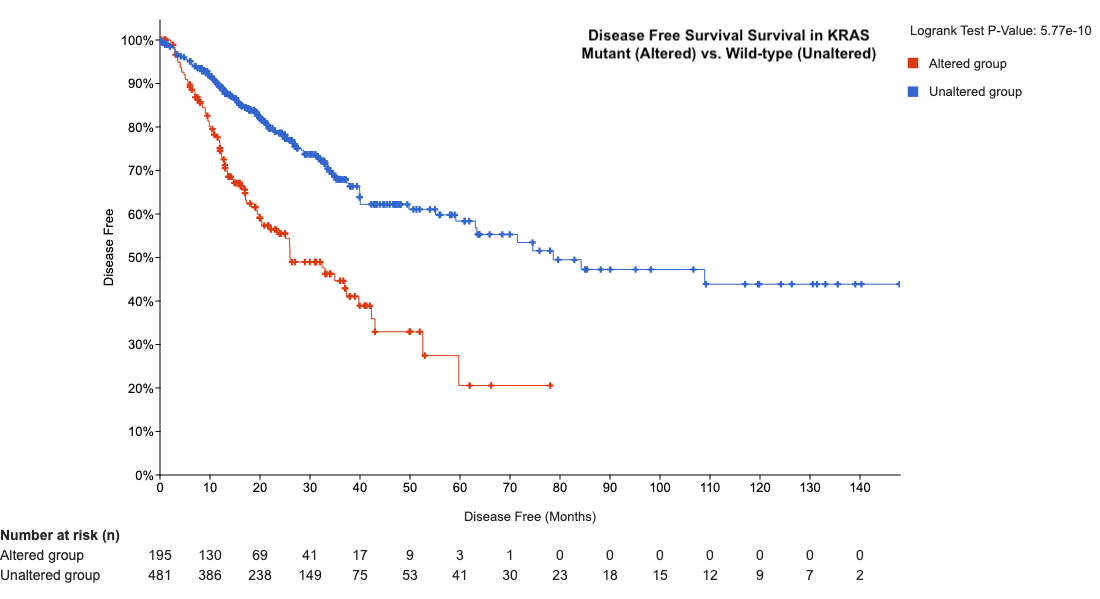
- Explore disease-free survival → Patients with mutant KRAS have lower disease-free survival.
More Ways to Use cBioPortal
- 🔬 Pathway analysis: Explore how mutations impact KRAS-related signaling pathways.
- 🧬 Copy number analysis: Compare CNAs in KRAS mutant vs. wild-type tumors.
- 🧑🤝🧑 Custom cohort building: Focus on subsets (e.g., smokers with KRAS mutations).
- ⏳ Tumor evolution: Analyze multi-sample progression from the same patient.
- 🛡️ Immuno-genomics: Study KRAS links to immune response (e.g., mutational burden).